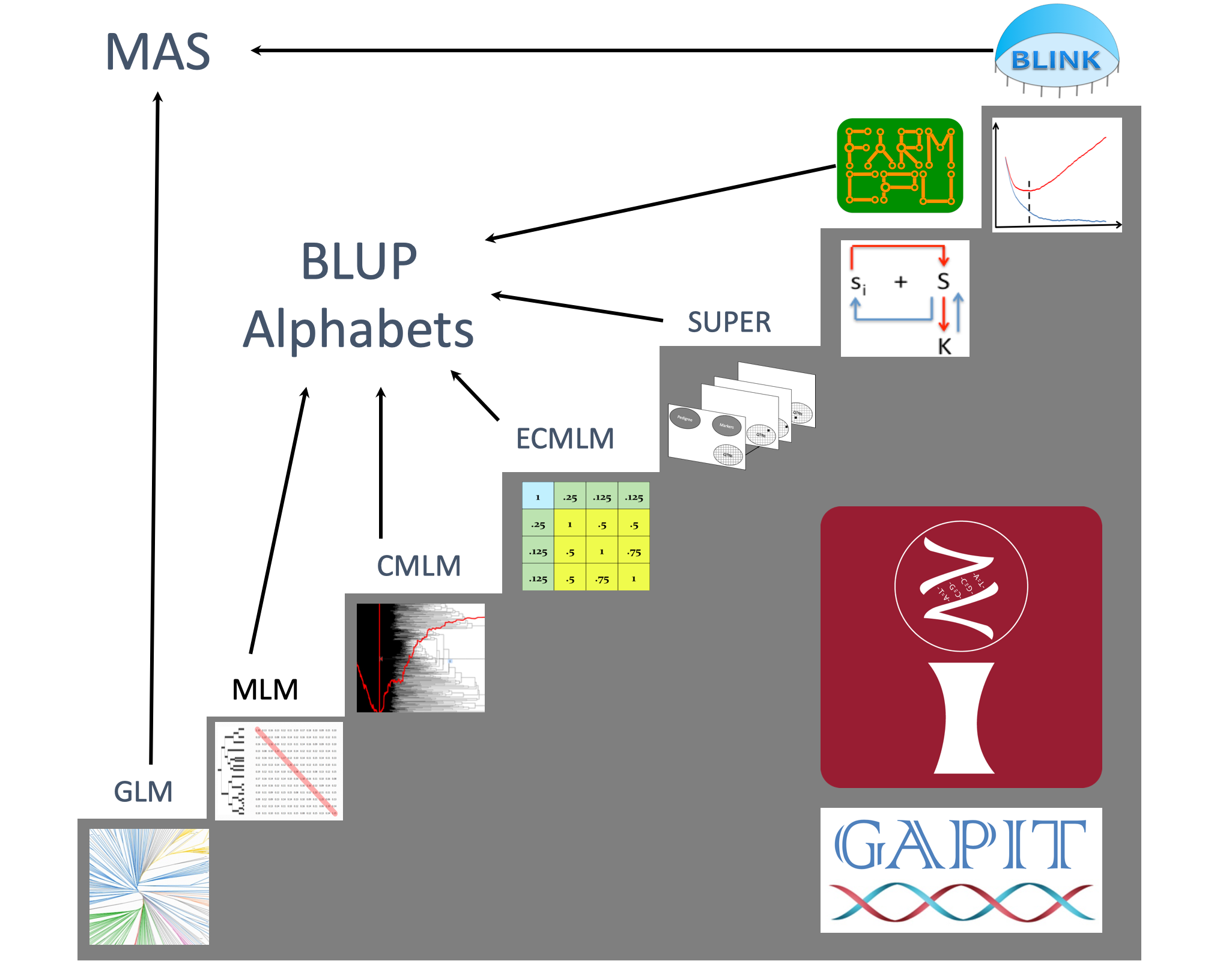
Genome Wide Association Study
As tests of linkage disequilibrium, Genome Wide Association Study (GWAS) was initiated by Spielman et al. in 1993 for dichotomous traits (Spielman et al. 1993) and by Allison in 1997 for quantitative traits (Allison 1997). To overcome false positives in this type of statistical tests, population structure was firstly incorporated, followed by kinship derived from all testing markers. To deal with the false negatives by the confounding between kinship and testing markers, the kinship is further optimized and eventually peeled completely using iterative multiple-locus models.
Phase 1: Simple tests using t, chi-square, and F statistics (Spielman et al. 1993
and Allison 1997);
Phase 2: Structure association tests using general linear model (Q matrix by Pritchard et al. 2000
and PCA by Price et al. 2006);
Phase 3: Structure and kinship association tests using mixed linear model (Q+K by Yu et al. 2005);
Phase 4: Optimized kinship (Compressed MLM by Zhang et al. 2010,
Enriched CMLM by Li et al 2014,
and SUPER GWAS by Wang et al 2014);
Phase 5: Peeled kinship: (MLMM by Segura et al. 2012,
FarmCPU by Liu et al. 2016,
and BLINK by Huang et al. 2019).
All the versions have been implemented to GAPIT with R Command Line Interface (CLI) and iPat with Graphic User Interface (GUI). As demonstrated by the background image below, the comparison of these methods can be conducted within 10 lines of R code. The optimized code (by ChatGPT) can be completed within a minute using parallel computation (ten times faster).

